IMPACT Tracks on the UCSC Genome Browser
IMPACT
We previously developed IMPACT, a machine learning model that predicts cell-type-specific regulatory element activity using master regulator transcription factors as anchors (Amariuta et al., 2019, American Journal of Human Genetics). IMPACT leverages more than five thousand epigenetic experiments across diverse cell types to describe the epigenetic patterns at the sites of actively bound transcription factors.
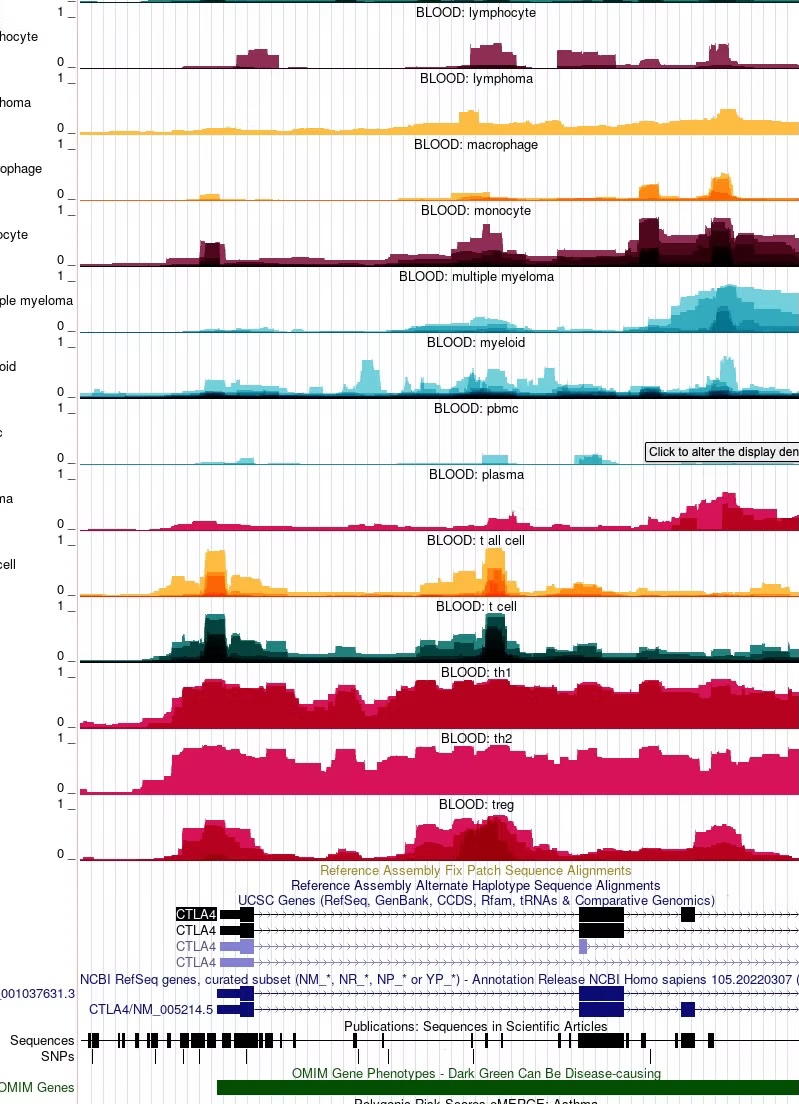
We have now computed the IMPACT regulatory element score across all ~3 billion base pairs of the genome for each of 707 unique transcription factor-cell type pairs. Using the UCSC genome browser, you can now query regions, genes, or DNA mutations of interest and visualize the predicted regulatory element activity across different cell types. This can reveal the cell types in which genes may be specifically expressed or the mechanisms that mutations are specifically perturbing.
For more information on the development of IMPACT and resulting predictive models, please navigate to the Tools Section of our website.
Viewing Transcription Factor-Specific Activity
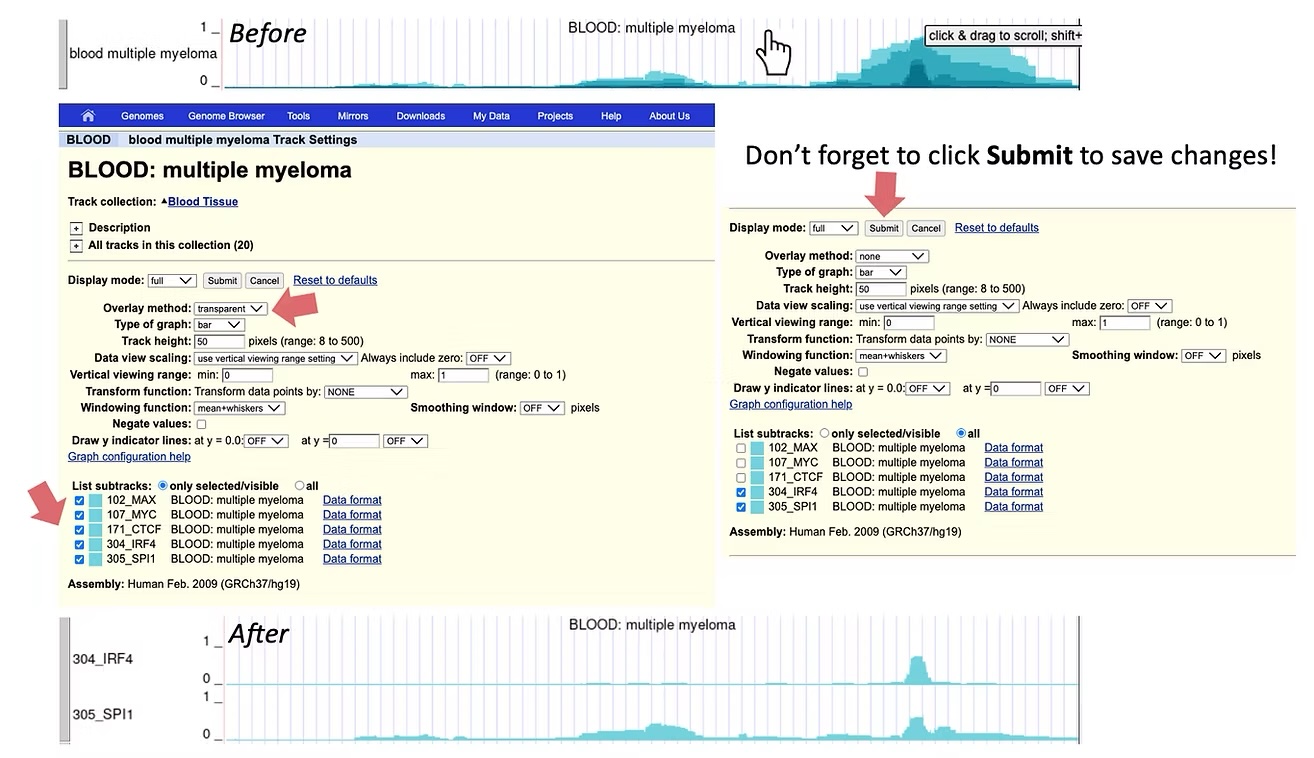
You may collapse or expand information across different IMPACT tracks from the same general tissue or cell type category to reveal consensus regulatory element activity (darker colors) compared to transcription factor-specific activity (lighter colors). By default IMPACT tracks are collapsed across models that share the same cell type, but are characterized by different transcription factors. To expand IMPACT tracks, click anywhere in the middle of the track, identify the Overlay Method option and select None from the drop-down menu. You may also select specific transcription factor tracks of interest by (de)-selecting the IMPACT tracks in List Substracts. The ID number for each track corresponds to the ID number assigned to the IMPACT track in the publication.
Data Format
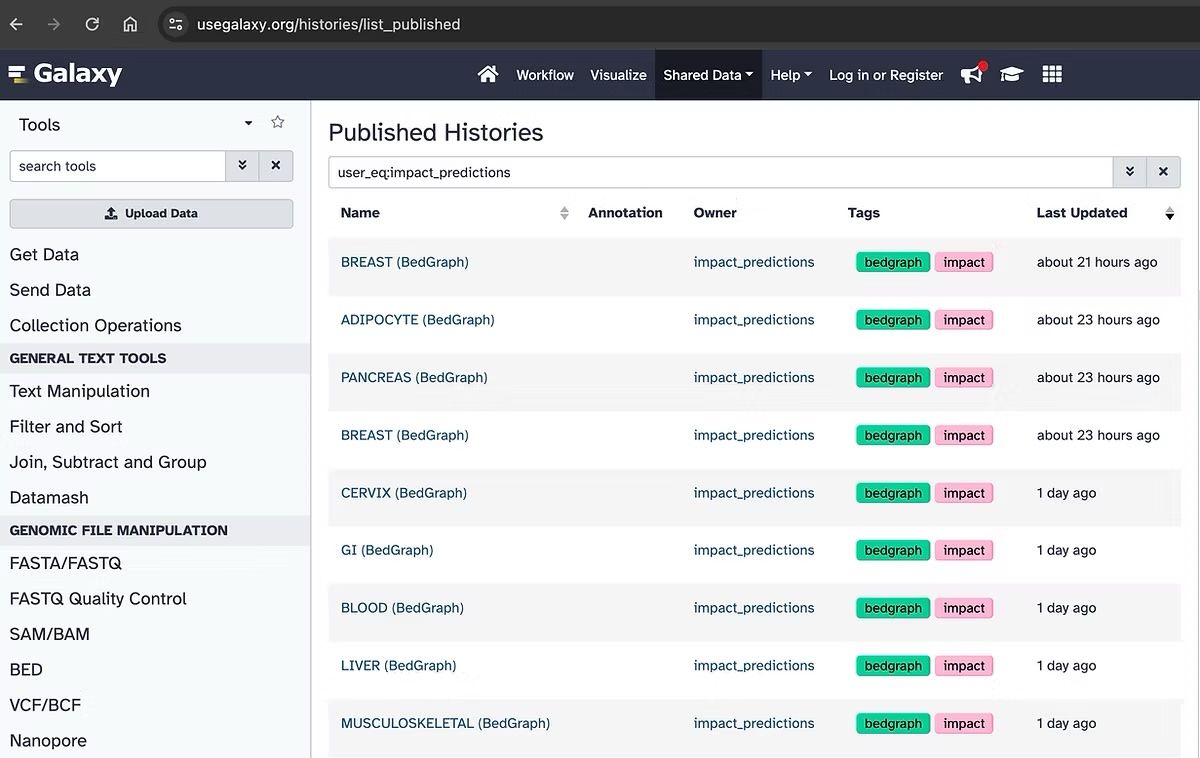
IMPACT prediction tracks are in .bigwig file format to be compatible with the UCSC genome browser. To download these bigwig files please visit our repository on Galaxy. While .bigwig files are binary files, we have also released IMPACT track predictions in .bedgraph file format for easier data manipulation and analysis. You can find all these data on galaxy, when searching user_eq:impact_predictions in Published Histories.
Acknowledgements
We thank Alona Korzhakova, a former Computational Data Science Researcher from our lab, and Kathryn Weinand from the Raychaudhuri Lab for generating genome-wide base pair resolution IMPACT tracks. We thank Alona for integrating them with the UCSC genome browser, such that these data now have full and direct access to the genomics community.
We would love to hear your feedback!
For questions or comments, please contact us at tamariutabartell [at] ucsd [dot] edu.